Unravelling Pain R&D Part 2: What Can Network Biology Teach Us?
In part one of this series, we delved into how open-source data sets, although vastly under-utilized, present an untapped resource that could potentially shift how we approach pain research and development.
In this part two, we will explore how the explosion of biomedical data will require complex integration and analysis to better understand the molecular basis of phenotypes.
Network Biology: A New Lens for Pain Research
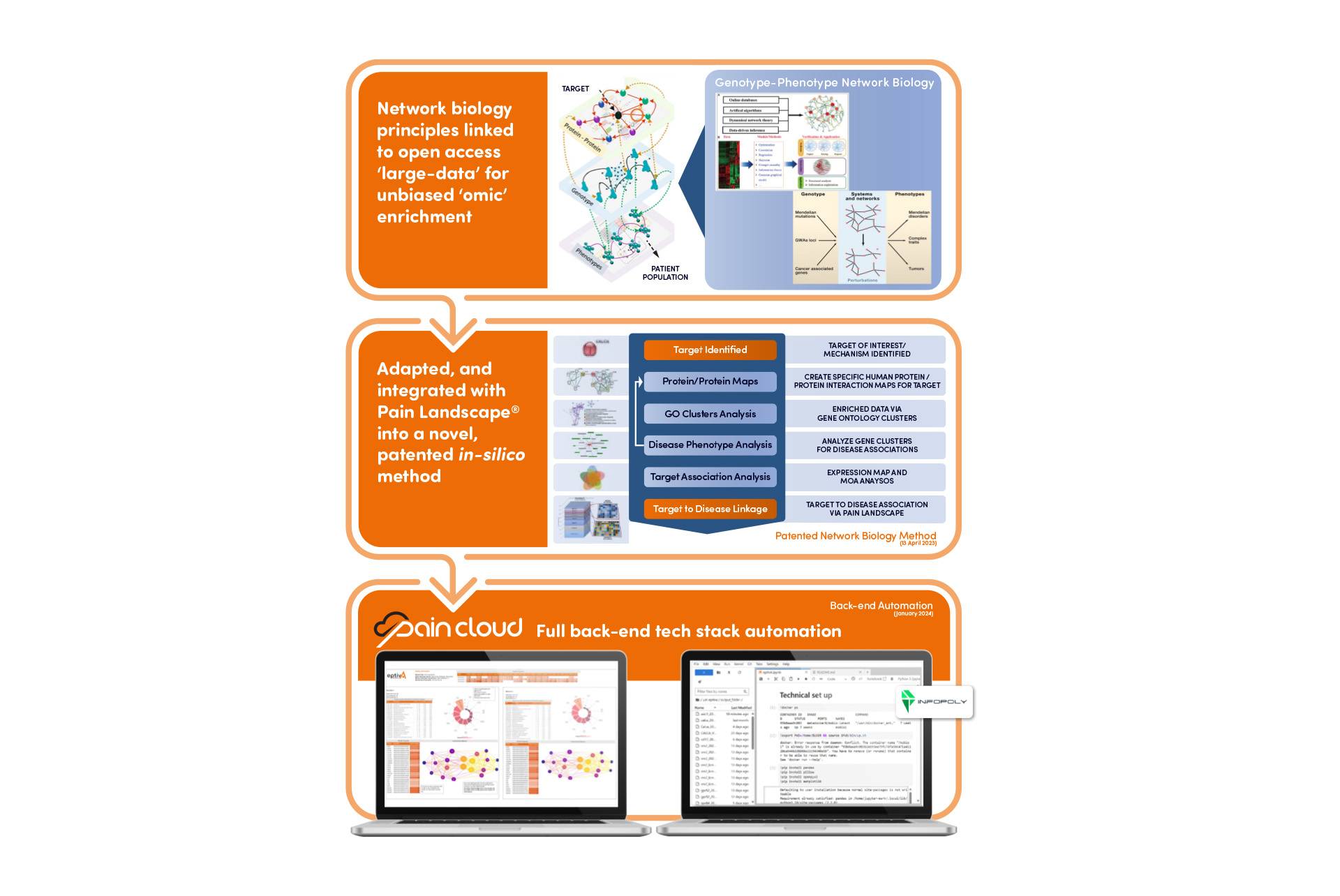
Network biology is a revolutionary approach that views biological systems not as isolated components, but as interconnected networks. Rather than focusing on single genes or proteins, network biology examines the complex interactions between multiple biological entities.
This approach represents a significant shift from traditional reductionist methods. Instead of breaking systems down into their smallest parts, network biology seeks to understand how these parts work together to create complex behaviours – like the experience of pain.
The concept of network biology has been gaining traction in medical research since the early 2000s, driven by advances in high-throughput ‘omic’ technologies and computational power. Its application to pain research, however, is opening up exciting new possibilities for understanding and treating this complex condition.
These stem from various factors, notably the growing complexity and volume of data together with the increased diversity of data types describing different tiers of biological organization.
The Power of Connections: From Proteins to Patients
Currently, much data exists in silos and is not analyzed in frameworks where all data are brought to bear in the development of biomarkers and novel functional targets. Network biology approaches, which emphasize the interactions between genes, proteins and metabolites, provide a framework for data integration such that genome, proteome, metabolome and other -omics data can be jointly analyzed to understand and predict disease phenotypes.
The ability to map protein-to-protein interactions is more than just an academic exercise – it has profound implications for our understanding of pain mechanisms and the development of new treatments.
By visualizing these complex networks, researchers can identify key nodes or hubs that play crucial roles in pain signalling. These could represent promising targets for new pain medications. Moreover, by comparing network patterns across different pain conditions, we might uncover shared mechanisms that could lead to more broadly effective treatments.
By understanding how slight variations in these protein networks can lead to different pain experiences, we move closer to tailoring treatments to individual patients – potentially explaining why two patients with seemingly identical conditions might respond differently to the same treatment.
Pain Cloud®: Mapping the Pain Universe
At the forefront of this revolution is Pain Cloud®, a groundbreaking proprietary database, build on the back of open-source data sets, that encompasses over 1,000 pain-related diseases.
This vast repository of information serves as a foundation for mapping protein-to-protein interactions on an unprecedented scale.
But what does this mean in practice? Essentially, Pain Cloud allows researchers to visualize the complex relationships between different proteins involved in pain pathways. It’s like having a detailed map of the pain universe, showing how different components interact and influence each other.
In 2022, we used the Pain Cloud platform to identify a novel link between cholecalciferol and specific pain conditions. This data was used to support a published method of use patent, which forms part of our IP and protection strategy for this new treatment approach.
We hope that this will provide a new choice of analgesic to patient groups with limited or no treatment options.
The Road Ahead: Embracing Network Biology and Open-Source Data
Recent research has illuminated the importance of tissue-specific regulatory networks and the pathways they encompass, which frequently manifest genetic mutations in particular patient cohorts.
This understanding emerged from the combined analysis of expression and chromatin accessibility data, unveiling a previously unidentified tissue-specific stem cell-like subtype of treatment-resistant prostate cancer that may be a target for intervention (Tang et al. 2022).1
Pain Cloud is part of an emerging frontier in systems and network approaches to medicine off the back of these advancements in the ability to model large-scale proteins interactions and how these are involved in the pain pathways of human systems.
We are among the first to be applying this methodology to pain research.
Our use of open-source data has paved the way to build a propriety IP that’s the first of its kind, which is enhancing network biology and helping pain medicine R&D benefit from:
- Accelerated discovery and innovation
- Enhanced collaboration and reducing duplication
- Improved reproducibility in research
- Democratized access to medical knowledge
By providing a more holistic view of pain mechanisms and enabling global collaboration, these approaches could accelerate the development of new treatments and improve outcomes for millions of pain sufferers.
The journey ahead is exciting, filled with the potential for groundbreaking discoveries and improved patient outcomes.