Unravelling Pain R&D Part 1: Leveraging Open-Source Databases
Open-source databases are collections of data that are freely available for anyone to use and enhance their own research..
This concept has been used for many years across several industries; one standout example of this is its use in the software industry. For example, NVIDIA has become one of the most successful businesses in history with its state-of-the-art graphics processing units (GPUs), all achieved whilst fully supporting open-source projects and releasing their own line of open-source GPUs.
Using open-source data can result in greater efficiency, organization, speed and success of development. Although this approach is rarely used in the healthcare industry, there is a huge wealth of high quality data becoming available.
In the 1990s, the Human Genome Project completely changed the landscape of open-source data being used in medicine, but the wider industry is still yet to fully leverage these databases for the benefit of research and development.
Here, we explore the advantages of utilizing and sharing open-source databases in pain and wider medical research and development.
We also share how Pain Cloud®, our novel digital B2B solution designed to aid the success of analgesic clinical development, can be a useful starting point in adopting this approach.
How is open-source data applicable to pain research?
The truth is that there is a huge amount of data already available to pain researchers that many aren’t using to their advantage. The use of these databases is fairly common in early chemistry, but it is yet to be adopted across the full spectrum of R&D activities.
Current methods stick closely to closed data models in which research is proprietary and access is restricted.
However, open-source data, often consisting of genomic sequences, clinical trial results and anonymized patient data, presents plenty of benefits for pain research which are simply under-utilized.
These benefits include:
- Accelerating discovery and innovation
With access to vast amounts of reliable open-source data, we can allow researchers to screen existing compounds for new therapeutic uses. This means that the research process, which is traditionally very time-intensive, can be drastically sped up.
This data can equip researchers to screen existing compounds for new therapeutic uses.
Open-source data can also be used to enhance proprietary research, contributing to researchers’ own models and solutions, paving the way for a collaborative landscape whilst maintaining a competitive approach.
- Prompting collaboration
Using open-source data allows researchers worldwide to build upon each other’s work, fostering a more collaborative medical research environment and minimizing the attrition rate of new drugs.
- Improving reproducibility
With access to dedicated open-source databases, researchers are able to build on each other’s research and verify and reproduce study results, resulting in reliable scientific findings.
Examining notable open-source databases in medicine
This international scientific research project set out to determine the base pairs that make up human DNA.
The project research conducted in an international collaboration for over two decades successfully identified, stored and shared access to the vast genetic content in chromosomes of the human genome.
Led by the National Institute of Health (NIH) and the U.S. Department of Energy (DOE), this project ran from 1990 to 2003, with the commitment to make all data available open-source for researchers across the globe. This has now led to over 1,600 molecular biology databases being available for open use, in part thanks to this innovative project.
The European Molecular Biology Laboratory (EMBL) was established in 1974 and has become a leader in biological research and making their data available through open-source databases.
EMBL is an intergovernmental organization with more than 80 independent research groups covering the spectrum of molecular biology
Those high-quality databases have been created and maintained by a consortia of leading research organizations with publications in multiple renowned scientific journals.
EMBL maintains one of the world’s most comprehensive ranges of freely available, up-to-date molecular data resources, with over 150 open-source databases and tools available.
This AI system, developed by Google DeepMind in co-operation with EMBL, predicts how a protein will fold by analyzing its amino acid sequence.
Identifying the molecular structure of proteins allows you to find chemicals that bind these proteins that can interact with them and become medicines.
However, once in situ, they fold and create their own environment as a protein. AlphaFold has revolutionized the way researchers can predict and prepare for this.
This is an example of an open-source database which is being used by many companies to enhance their own proprietary research in early chemistry.
How can we enhance R&D activities by taking advantage of high-quality open source databases?
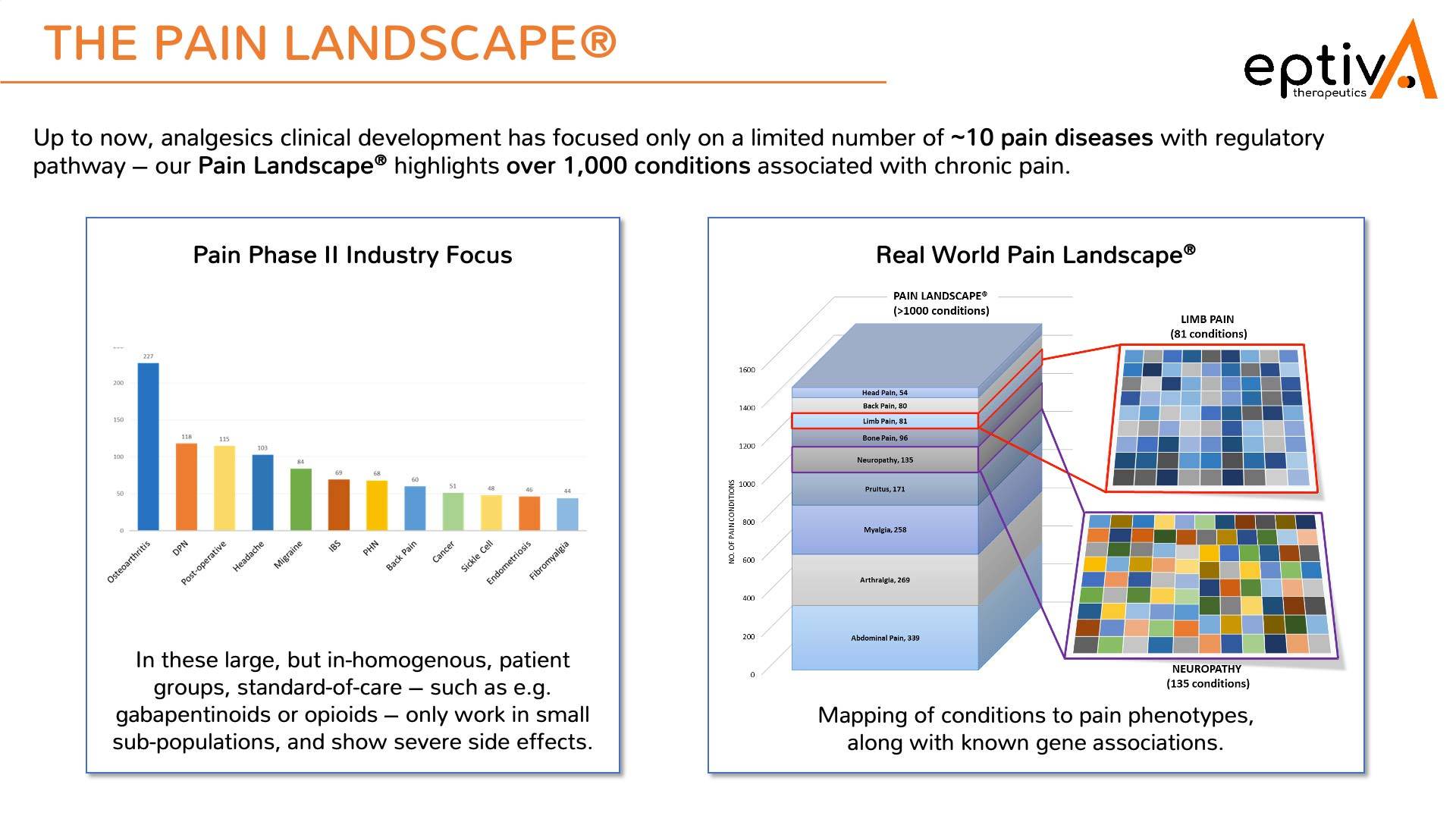
Pain Cloud® is an example of how it is possible to link multiple open-source data to in-house expertise and create a novel method to enhance R&D. Our proprietary Pain Landscape® was created over a number of years to provide a database of over one thousand pain conditions . However, on the front-end of the platform, we used multiple well-established, high-quality open-source databases which link together before linking to the Pain Landscape.
Designed in partnership with technology company Infopoly Ltd., Pain Cloud has become an automated platform with the development of a back-end tech stack via integrated novel software.
The network biology algorithm can now be operated, automatically accessing large data sets from high-quality molecular databases, providing unbiased analysis linking research targets to optimized disease phenotypes.
The cloud-based digitization enables fast, precise results with novel software applications providing proprietary data visualizations.
We believe Pain Cloud is one of the first platforms using an in-silico approach, stringing databases together in a novel manner.
This leads to a greater probability of success, allowing your compound to reach the right patient group and, in turn, your medicine to the right patient.
The use of open-source data ties in closely with network biology, a revolutionary approach to medicine that views biological systems not just as isolated components, but as interconnected networks.
If you have any questions, feel free to get in touch.